Motivation
Many studies indicate that long non coding RNAs (lncRNAs) carry out very diverse biological functions and play critical roles in various kinds of diseases. Identitying and discovering new lncRNA transcripts have been the fundamental process in lncRNA-related research. Currently, sequencing technologies provides us with thousands of novel transcripts, which demands a more accurate and effective algorithm to perform lncRNA identification.Results
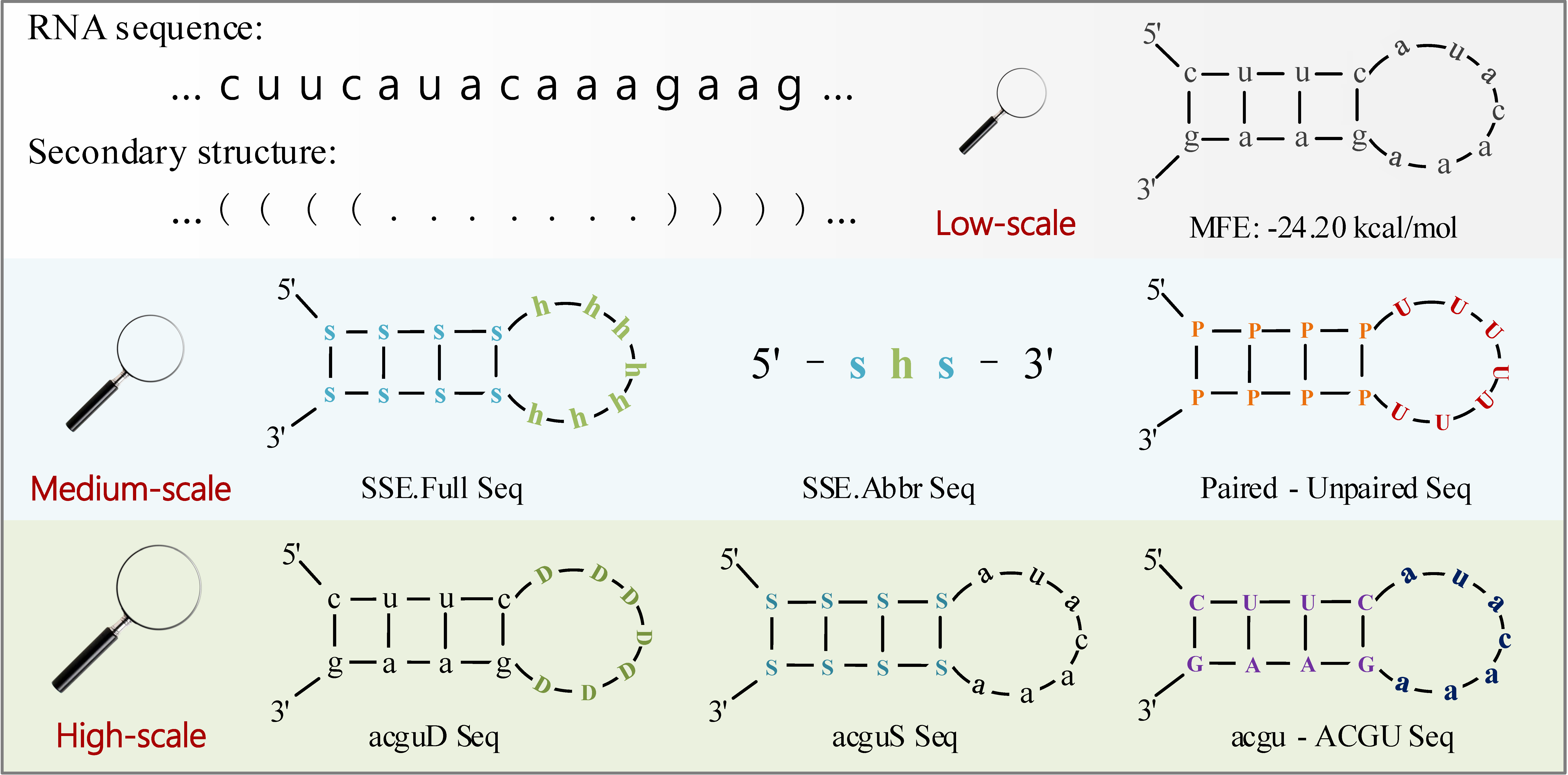
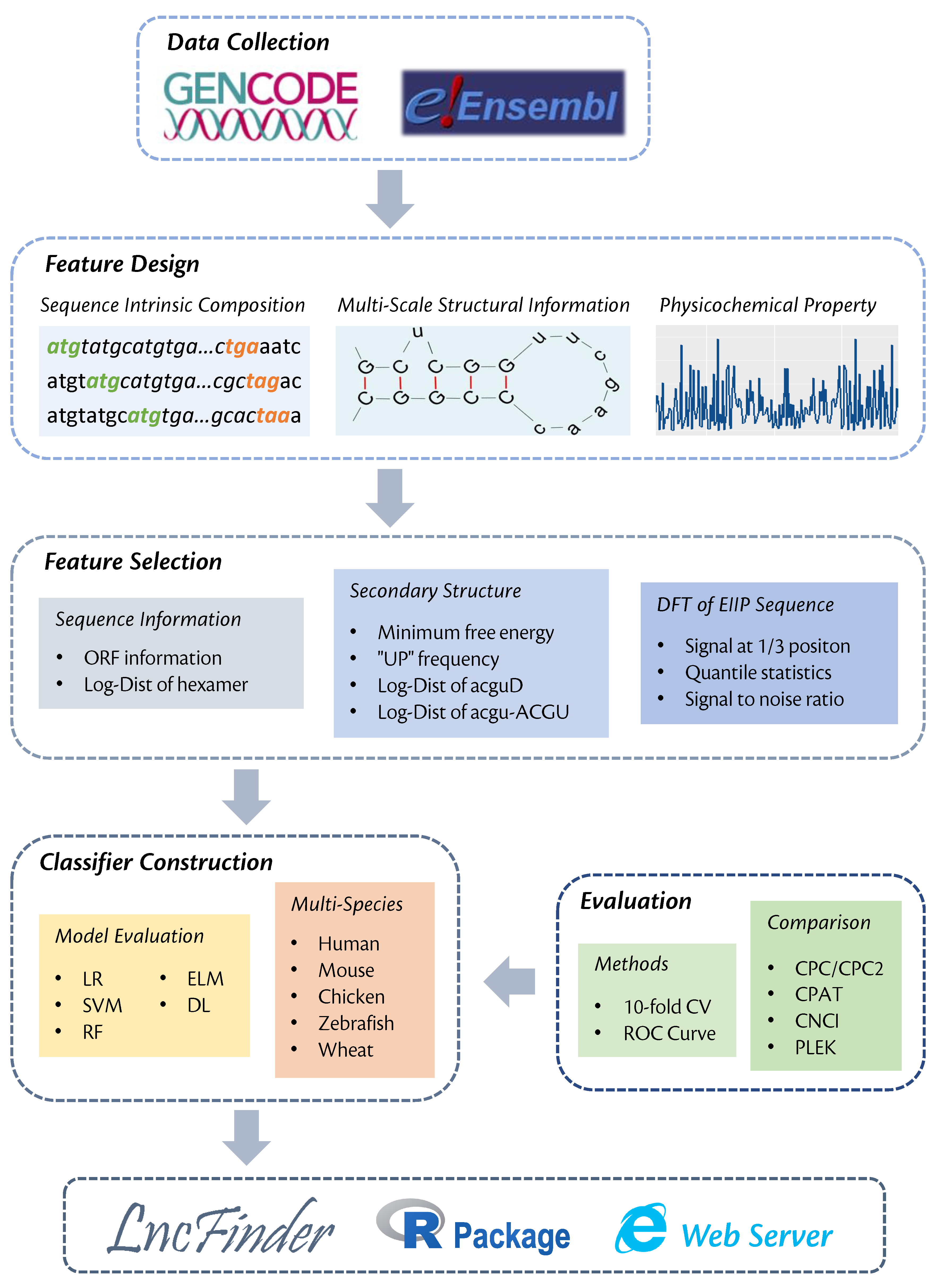
A new lncRNA identification tool, LncFinder, is developed based on Logarithm-Distance of hexamer, multi-scale structural information and physicochemical features obtained from Fast Discrete Fourier Transforms. In order to determine the optimal classifier, five widely used machine learning algorithms: logistic regression, support vector machine (SVM), random forest, extreme learning machine and deep learning are validated using 10-fold cross validation. SVM is finally selected as the classifier of LncFinder. Having been evaluated with comprehensive feature selection and model validation schemes, LncFinder outperforms several state-of-the-art tools on multiple species. Users can re-train LncFinder with new datasets or different machine learning algorithms easily and efficiently. Standalone version of LncFinder is released as R package, and a web server is also developed to maximise its availability. R package can be downloaded from CRAN: https://CRAN.R-project.org/package=LncFinder.Illustration of Multi-scale Secondary Structural Sequences
Construction of LncFinder
Copyright © 2017-2018. All Rights Reserved. Developed by HAN Siyu & XU Yangyi, Machine Learning Group @ Jilin University.